Alternative RNA Splicing
Overview

Alternative RNA splicing treats different segments of mRNA as exons depending on the snRNA on the spliceosome. This allows for multiple proteins to be coded from the same mRNA strand
Spliceosome
The spliceosome's six complexes recognize the splice sites of the exon and cut and bind them together.
The U1 and U2 snRNPs identify the edges of the exon by recognizing the 5' and 3' splice sites with their snRNA. They do this by binding to the respective GU (5') or AG (3') sequences present on the edges of 99% of exons.
The remaining U3, U4, U5, and U6 snRNPs are used to cut out the intron and connect the exons together.
snRNA
snRNA is RNA that makes up part of the spliceosome. Its function as an RNA is imperative to the splicing process because it can recognize the splice sites by matching nucleotide sequences.
vs
snRNP
snRNP is a combination of snRNA and functional protein groups. The snRNP recognizes the splice sites and cuts the mRNA strand. A total of 6 snRNPs are used in the splicing process.
Examples
Fas Receptor
The Fas receptor promotes apoptosis -- the programmatic killing of cells -- for cancer cells. When Exon 6 is excluded, the Fas receptor becomes soluble and dissolves, making it unable to promote apoptosis.
TIA-1 and PTB are two antagonistic proteins that affect how the snRNP binds to the exon.
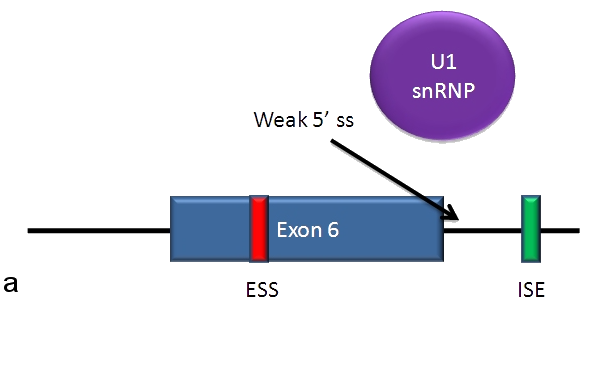
Without either TIA-1 or PTB, the U1 complex of the spliceosome has a weak connection to the 5' side of the exon. This results in the exon being skipped and the soluble Fas receptor being produced.
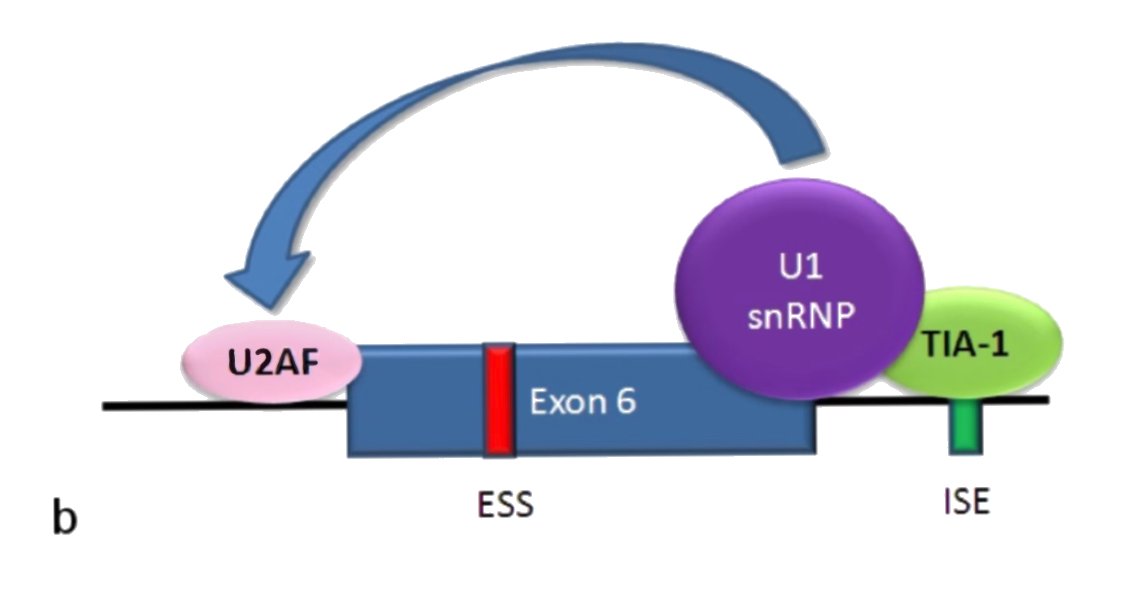
When TIA-1 is present, the U1 complex of the spliceosome has a strong connection to the 5' side of the exon. Additionally, the 5' donor site assists the U2AF in binding to the 3' side of the exon. This results in the exon being included and the membrane-bound Fas receptor being produced.
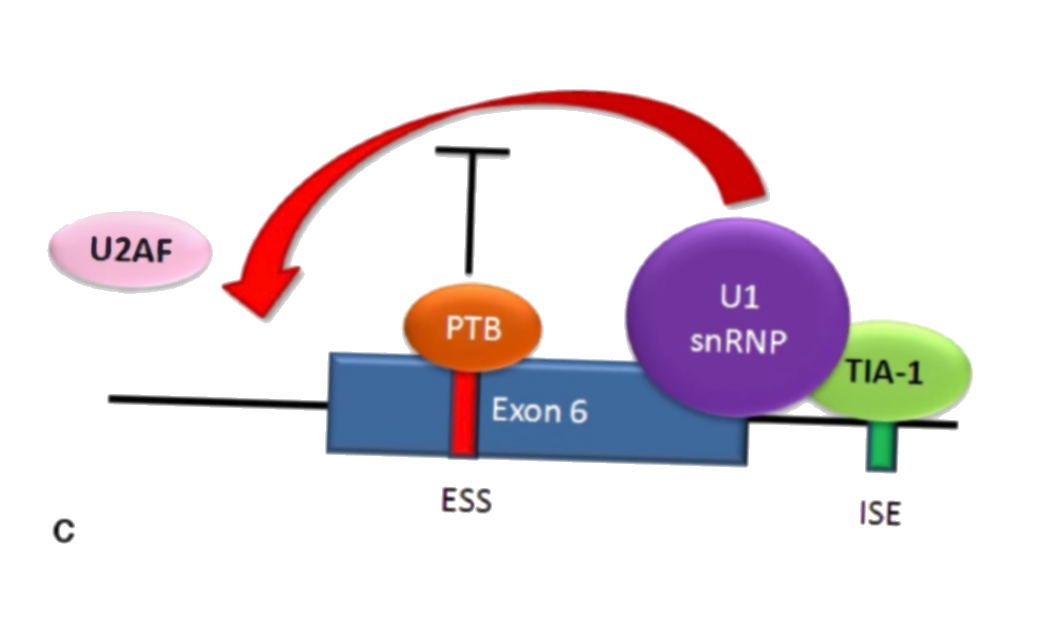
When both TIA-1 and PTB are present, the PTB inhibits the 5' donor complex. This results in the U2AF being unable to bind to the 3' side of the exon, and the exon being skipped.
drosophila dsx
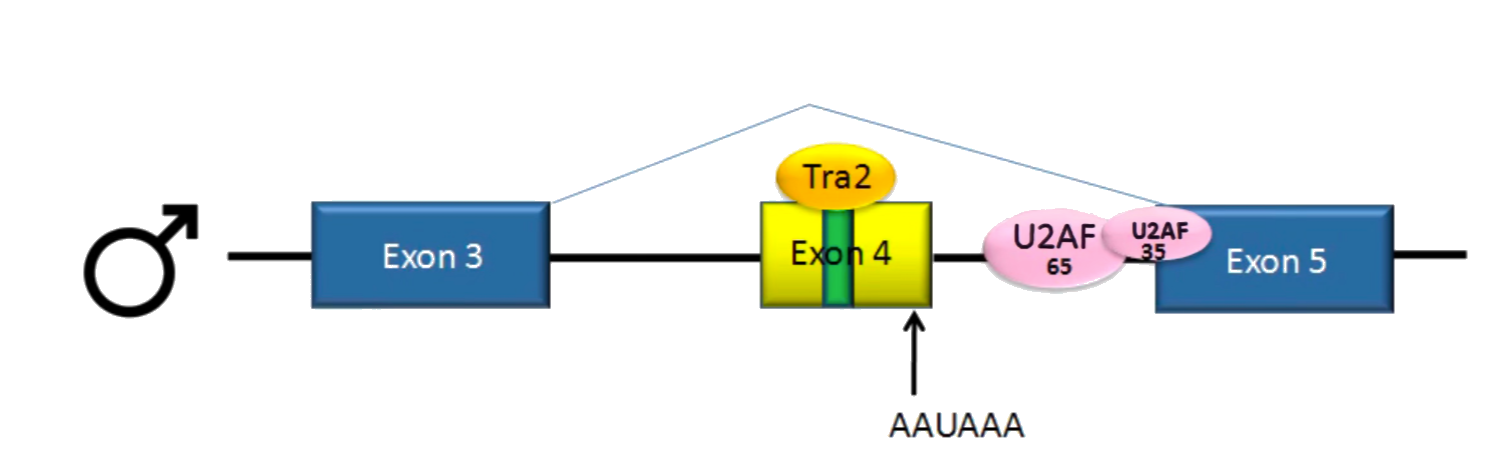
Why does exon 4 get cut off for male flies?
- No splicing activator
- during splicing, the polypyrimidine tract acts as a signal for the splicesome to recognize it so that it can cut from the correct SS
- Exon 4 has a weak polypyrimidine tract (meaning it doesn't match the nucleotide sequence on the SS well) → the U2AF doesn't bind well
- U2AF → protein that recognizes specific sequence in the polypyrimidine tract + recruits and accurately positions snRNP to the correct site
- When U2AF doesn't bind well → 3' SS on that exon is not cut

Why is exon 4 included for female flies?
- Females produce splicing activator → Tra
- If Tra is present in exon 4, it binds to Tra2 with another SR protein (certain fam of protein)
- Protein Tra2 is produced in both sexes + binds to exonic splicing enhancers (ESE = sequences of nucleotides in pre-mRNA that enhance the efficiency of splicing) in exon
- forms a complex that helps U2AF bind to weak polypyrimidine tract ⇒ splicing is normal